Examples
Create your own sound
Steps
- Install and start Processing
- Copy code of next section into processing
- Modify file name and – via that – the input file (ATTENTION: exact file name format is important!)
- Run program and enjoy!
Code
// Recommended to use Processing 3.5.4 (V4 and above have no mididbus support)
// Save file in format "sketch_yymmdd_abcd" with "abcd" being a four letter rcsb code to specify molecule (e.g. "1HOC"), save, and run
import themidibus.*;
import java.io.File;
MidiBus myBus;
Table table;
float[] xVal, yVal, zVal,x,y;
int[] xscaled, yscaled, zscaled;
float[] xoriginal, yoriginal, zoriginal;
float xmin, xmax, ymin, ymax, zmin, zmax, wert1, wert2, wert3;
int len;
int[] tone;
int i = 0;
int j = 0;
float border = 0.1;
float dj;
float[] p1a, p1b, p2a, p2b, p3a, p3b;
void setup() {
size(1000, 1000);
background(0);
String scriptPath = System.getProperty("user.dir");
File currentScript = new File(scriptPath + File.separator + this.getClass().getSimpleName() + ".pde");
String myString = (currentScript.getName());
String protein = myString.substring(14,18);
println(protein);
myBus = new MidiBus(this, "Real Time Sequencer", "Microsoft MIDI Mapper");
saveData(protein);
scaleData(protein);
}
void saveData(String protein) {
String filename = "https://files.rcsb.org/view/" + protein + ".cif";
String[] lines = loadStrings(filename);
ArrayList<String> xvalList = new ArrayList<String>();
ArrayList<String> yvalList = new ArrayList<String>();
ArrayList<String> zvalList = new ArrayList<String>();
for (String mainString : lines) {
String[] words = mainString.split("\\s+");
if (words.length > 0 && words[0].equals("ATOM") && words[3].equals("CA")) {
xvalList.add(words[10]);
yvalList.add(words[11]);
zvalList.add(words[12]);
}
}
ArrayList<String> csvContentList = new ArrayList<String>();
csvContentList.add(join(new String[]{"x", "y", "z"}, ","));
for (int k = 0; k < xvalList.size(); k++) {
csvContentList.add(join(new String[]{xvalList.get(k), yvalList.get(k), zvalList.get(k)}, ","));
}
String outputFilename = "output_" + protein + ".csv";
saveStrings(outputFilename, csvContentList.toArray(new String[0]));
}
void scaleData(String protein) {
table = loadTable("output_" + protein + ".csv", "header");
len = table.getRowCount();
// Initialize arrays
xVal = new float[len];
yVal = new float[len];
zVal = new float[len];
p1a = new float[len];
p1b = new float[len];
p2a = new float[len];
p2b = new float[len];
p3a = new float[len];
p3b = new float[len];
for (int row = 0; row < len; row++) {
xVal[row] = table.getFloat(row, "x");
yVal[row] = table.getFloat(row, "y");
zVal[row] = table.getFloat(row, "z");
}
xmin = min(xVal);
xmax = max(xVal);
ymin = min(yVal);
ymax = max(yVal);
zmin = min(zVal);
zmax = max(zVal);
for (int row = 0; row < len; row++) {
wert1 = map(table.getFloat(row, "x"), xmin, xmax, 0,height/2);
wert2 = map(table.getFloat(row, "y"), ymin, ymax, 0,height/2);
wert3 = map(table.getFloat(row, "z"), zmin, zmax, 0,height/2);
xVal[row] = wert1;
yVal[row] = wert2;
zVal[row] = wert3;
}
dj = TWO_PI / len;
initializePoints(xVal, p1a, p1b);
initializePoints(yVal, p2a, p2b);
initializePoints(zVal, p3a, p3b);
}
void draw() {
if (i < len - 1) {
drawSequence(xVal, p1a, p1b, color(255,255,255));
drawSequence(yVal, p2a, p2b, color(255,127,36));
drawSequence(zVal, p3a, p3b, color(139,90,43));
i++;
} else if (i == len - 1) {
// Draw a closing line back to zero or to a specified point
drawClosingLines(xVal, p1a, p1b, color(255,255,255));
drawClosingLines(yVal, p2a, p2b, color(255,127,36));
drawClosingLines(zVal, p3a, p3b, color(139,90,43));
i++; // Increment to avoid re-entering this block
}
if (mousePressed == true) { saveFrame("pic.png");};
saveFrame("picfinal.png");
}
void initializePoints(float[] sequence, float[] x, float[] y) {
for (int j = 0; j < len; j++) {
x[j] = width/2 + sequence[j] * cos(j * dj);
y[j] = width/2 + sequence[j] * sin(j * dj);
}
}
void drawClosingLines(float[] sequence, float[] x, float[] y, color strokeColor) {
stroke(strokeColor);
line(x[len - 1], y[len - 1], x[0], y[0]); // Connect the last point back to center or origin
}
void drawSequence(float[] sequence, float[] x, float[] y, color strokeColor) {
int tone = (int) sequence[i];
myBus.sendNoteOn(1, tone*127/width*2 , tone);
float r = random(1, 3);
delay(int(r) * 80);
stroke(strokeColor);
x[i] = width/2 + sequence[i] * cos(i * dj);
y[i] = width/2 + sequence[i] * sin(i * dj);
line(x[i], y[i], x[i+1], y[i+1]);
}
How the sounds were made
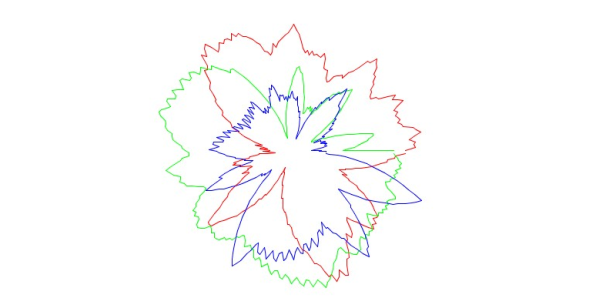
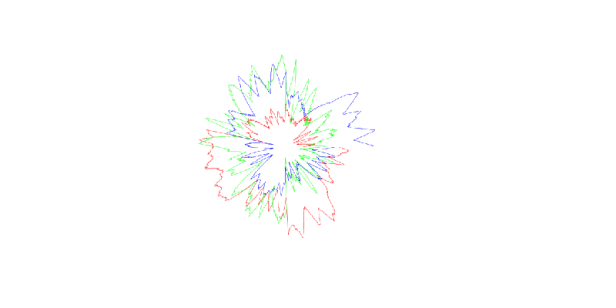
During an artist residency at the Springer Lab at Constructor University Bremen – exploring biomolecules such as proteins and viruses in various contexts – a computer program was developed to transform atomic positions of biomolecules into MIDI sounds while simultaneously visualizing the conversion through flower-like polar coordinate shapes.
The input data consisted of the x, y, and z coordinates of all atoms of a specified molecule, retrieved from the molecular database RCSB PDB. These coordinates generated both a three-voice sound and a visualization looking like circular canons.
That way, each biomolecule produces a unique sound and shape, inviting unconventional perspectives to compare, celebrate, and appreciate nature’s endlessly varied yet intrinsically related structures.